

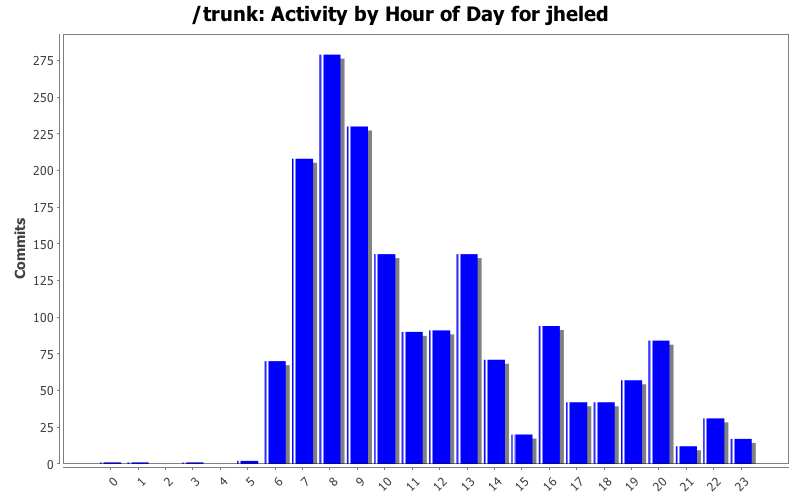
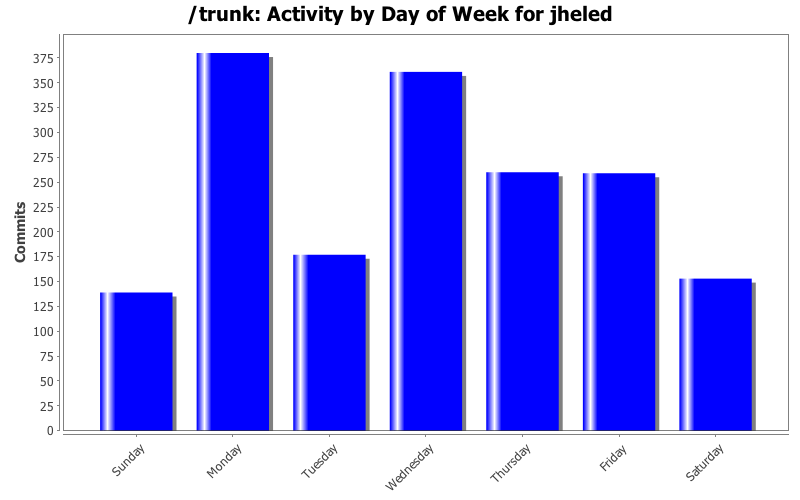
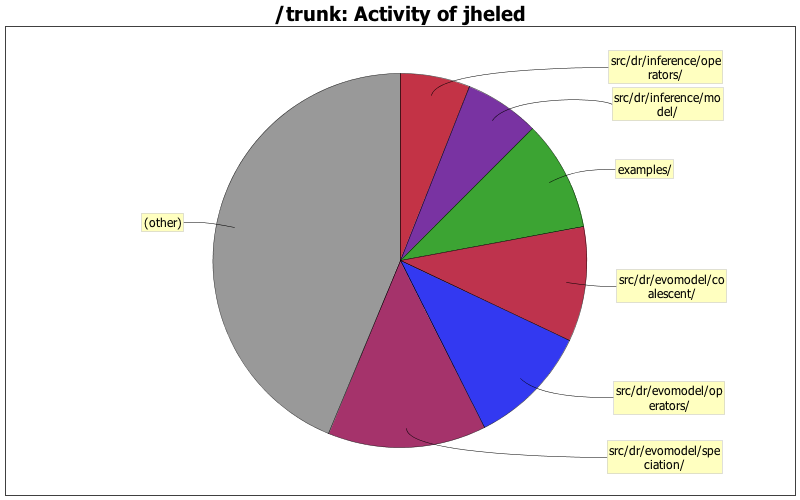
| Directory | Changes | Lines of Code | Lines per Change |
|---|---|---|---|
| Totals | 1729 (100.0%) | 36249 (100.0%) | 20.9 |
| src/dr/evomodel/speciation/ | 98 (5.7%) | 4948 (13.7%) | 50.4 |
| src/dr/evomodel/operators/ | 60 (3.5%) | 3817 (10.5%) | 63.6 |
| src/dr/evomodel/coalescent/ | 126 (7.3%) | 3597 (9.9%) | 28.5 |
| examples/ | 23 (1.3%) | 3473 (9.6%) | 151.0 |
| src/dr/inference/model/ | 126 (7.3%) | 2349 (6.5%) | 18.6 |
| src/dr/inference/operators/ | 78 (4.5%) | 2190 (6.0%) | 28.0 |
| src/dr/inference/trace/ | 28 (1.6%) | 1262 (3.5%) | 45.0 |
| src/dr/app/beauti/ | 45 (2.6%) | 902 (2.5%) | 20.0 |
| src/dr/xml/ | 72 (4.2%) | 873 (2.4%) | 12.1 |
| examples/release/calibrations/ | 19 (1.1%) | 754 (2.1%) | 39.6 |
| src/dr/evomodel/tree/ | 58 (3.4%) | 750 (2.1%) | 12.9 |
| src/dr/evolution/tree/ | 51 (2.9%) | 733 (2.0%) | 14.3 |
| src/dr/app/beauti/generator/ | 37 (2.1%) | 706 (1.9%) | 19.0 |
| src/dr/inference/loggers/ | 23 (1.3%) | 661 (1.8%) | 28.7 |
| src/dr/evolution/coalescent/ | 71 (4.1%) | 661 (1.8%) | 9.3 |
| src/dr/evomodel/substmodel/ | 46 (2.7%) | 654 (1.8%) | 14.2 |
| src/dr/inference/distribution/ | 39 (2.3%) | 520 (1.4%) | 13.3 |
| src/dr/app/tools/ | 29 (1.7%) | 497 (1.4%) | 17.1 |
| src/dr/inference/markovchain/ | 15 (0.9%) | 481 (1.3%) | 32.0 |
| src/dr/inference/prior/ | 26 (1.5%) | 471 (1.3%) | 18.1 |
| src/dr/app/oldbeauti/ | 12 (0.7%) | 417 (1.2%) | 34.7 |
| src/dr/evomodel/continuous/ | 22 (1.3%) | 388 (1.1%) | 17.6 |
| examples/treeOpsTest/ | 3 (0.2%) | 302 (0.8%) | 100.6 |
| src/test/dr/evomodel/substmodel/ | 7 (0.4%) | 282 (0.8%) | 40.2 |
| src/dr/app/util/ | 6 (0.3%) | 261 (0.7%) | 43.5 |
| src/dr/math/distributions/ | 12 (0.7%) | 257 (0.7%) | 21.4 |
| src/dr/util/ | 32 (1.9%) | 213 (0.6%) | 6.6 |
| src/dr/exporters/ | 4 (0.2%) | 208 (0.6%) | 52.0 |
| src/test/dr/distibutions/ | 4 (0.2%) | 202 (0.6%) | 50.5 |
| src/dr/app/beast/ | 32 (1.9%) | 172 (0.5%) | 5.3 |
| src/dr/evomodel/coalescent/operators/ | 9 (0.5%) | 161 (0.4%) | 17.8 |
| src/dr/java16compat/ | 3 (0.2%) | 160 (0.4%) | 53.3 |
| src/dr/evomodelxml/speciation/ | 7 (0.4%) | 156 (0.4%) | 22.2 |
| src/dr/evolution/util/ | 14 (0.8%) | 155 (0.4%) | 11.0 |
| src/dr/evomodel/treelikelihood/ | 25 (1.4%) | 146 (0.4%) | 5.8 |
| src/dr/evomodelxml/ | 21 (1.2%) | 137 (0.4%) | 6.5 |
| src/dr/evolution/io/ | 10 (0.6%) | 133 (0.4%) | 13.3 |
| src/dr/inference/mcmc/ | 15 (0.9%) | 101 (0.3%) | 6.7 |
| src/dr/evoxml/ | 37 (2.1%) | 100 (0.3%) | 2.7 |
| src/dr/gui/chart/ | 8 (0.5%) | 95 (0.3%) | 11.8 |
| src/dr/evomodel/arg/ | 16 (0.9%) | 90 (0.2%) | 5.6 |
| src/dr/app/treestat/statistics/ | 14 (0.8%) | 88 (0.2%) | 6.2 |
| src/dr/math/ | 17 (1.0%) | 86 (0.2%) | 5.0 |
| src/dr/app/treestat/ | 7 (0.4%) | 86 (0.2%) | 12.2 |
| src/dr/app/tracer/traces/ | 12 (0.7%) | 85 (0.2%) | 7.0 |
| src/dr/app/tracer/application/ | 9 (0.5%) | 85 (0.2%) | 9.4 |
| src/dr/evomodel/sitemodel/ | 23 (1.3%) | 84 (0.2%) | 3.6 |
| src/dr/stats/ | 4 (0.2%) | 68 (0.2%) | 17.0 |
| src/dr/app/beagle/evomodel/substmodel/ | 4 (0.2%) | 62 (0.2%) | 15.5 |
| src/dr/evomodel/branchratemodel/ | 20 (1.2%) | 61 (0.2%) | 3.0 |
| src/dr/evomodel/indel/ | 15 (0.9%) | 59 (0.2%) | 3.9 |
| src/dr/evomodel/arg/operators/ | 12 (0.7%) | 58 (0.2%) | 4.8 |
| src/dr/evomodel/transmission/ | 13 (0.8%) | 57 (0.2%) | 4.3 |
| src/dr/app/beauti/components/ | 5 (0.3%) | 57 (0.2%) | 11.4 |
| src/dr/evomodelxml/tree/ | 9 (0.5%) | 52 (0.1%) | 5.7 |
| src/dr/app/beauti/options/ | 7 (0.4%) | 50 (0.1%) | 7.1 |
| src/test/dr/evomodel/coalescent/ | 3 (0.2%) | 48 (0.1%) | 16.0 |
| src/dr/app/beastdev/ | 8 (0.5%) | 46 (0.1%) | 5.7 |
| src/dr/evolution/alignment/ | 4 (0.2%) | 45 (0.1%) | 11.2 |
| src/dr/app/beagle/evomodel/parsers/ | 6 (0.3%) | 44 (0.1%) | 7.3 |
| src/dr/evolution/colouring/ | 6 (0.3%) | 41 (0.1%) | 6.8 |
| src/dr/evolution/wrightfisher/ | 6 (0.3%) | 35 (0.1%) | 5.8 |
| src/dr/app/plugin/ | 2 (0.1%) | 32 (0.1%) | 16.0 |
| src/dr/gui/table/ | 2 (0.1%) | 27 (0.1%) | 13.5 |
| src/test/dr/evomodel/speciation/ | 9 (0.5%) | 26 (0.1%) | 2.8 |
| src/dr/inference/ml/ | 4 (0.2%) | 24 (0.1%) | 6.0 |
| src/dr/evomodel/simulator/ | 2 (0.1%) | 24 (0.1%) | 12.0 |
| src/dr/app/beauti/util/ | 2 (0.1%) | 24 (0.1%) | 12.0 |
| src/dr/inferencexml/ | 6 (0.3%) | 20 (0.1%) | 3.3 |
| src/dr/evomodel/beagle/parsers/ | 8 (0.5%) | 20 (0.1%) | 2.5 |
| src/dr/evomodel/arg/coalescent/ | 6 (0.3%) | 20 (0.1%) | 3.3 |
| src/test/dr/evomodel/operators/ | 6 (0.3%) | 18 (0.0%) | 3.0 |
| src/dr/evomodel/coalescent/structure/ | 6 (0.3%) | 18 (0.0%) | 3.0 |
| src/dr/evomodel/MSSD/ | 9 (0.5%) | 18 (0.0%) | 2.0 |
| src/dr/app/coalgen/ | 4 (0.2%) | 18 (0.0%) | 4.5 |
| src/dr/evomodelxml/coalescent/ | 3 (0.2%) | 16 (0.0%) | 5.3 |
| src/dr/app/tracer/analysis/ | 2 (0.1%) | 16 (0.0%) | 8.0 |
| src/dr/gui/tree/ | 5 (0.3%) | 15 (0.0%) | 3.0 |
| src/dr/evomodel/clock/ | 10 (0.6%) | 14 (0.0%) | 1.4 |
| src/dr/inferencexml/operators/ | 4 (0.2%) | 13 (0.0%) | 3.2 |
| src/dr/app/beagle/evomodel/treelikelihood/ | 1 (0.1%) | 11 (0.0%) | 11.0 |
| src/dr/inference/mcmcmc/ | 3 (0.2%) | 10 (0.0%) | 3.3 |
| src/dr/evolution/datatype/ | 2 (0.1%) | 10 (0.0%) | 5.0 |
| src/dr/gui/graph/ | 4 (0.2%) | 9 (0.0%) | 2.2 |
| src/dr/evomodel/tree/randomlocalmodel/ | 3 (0.2%) | 9 (0.0%) | 3.0 |
| src/dr/geo/ | 1 (0.1%) | 8 (0.0%) | 8.0 |
| src/dr/evolution/distance/ | 2 (0.1%) | 8 (0.0%) | 4.0 |
| src/dr/inferencexml/model/ | 3 (0.2%) | 7 (0.0%) | 2.3 |
| src/dr/inferencexml/distribution/ | 2 (0.1%) | 7 (0.0%) | 3.5 |
| src/dr/evomodel/beagle/substmodel/ | 1 (0.1%) | 7 (0.0%) | 7.0 |
| src/dr/evolution/coalescent/structure/ | 2 (0.1%) | 7 (0.0%) | 3.5 |
| src/test/dr/evomodel/arg/operators/ | 2 (0.1%) | 5 (0.0%) | 2.5 |
| src/dr/math/functionEval/ | 3 (0.2%) | 5 (0.0%) | 1.6 |
| src/dr/evomodelxml/operators/ | 3 (0.2%) | 5 (0.0%) | 1.6 |
| src/dr/app/seqgen/ | 1 (0.1%) | 5 (0.0%) | 5.0 |
| src/dr/app/beauti/datapanel/ | 1 (0.1%) | 5 (0.0%) | 5.0 |
| src/dr/matrix/ | 4 (0.2%) | 4 (0.0%) | 1.0 |
| src/dr/evomodel/newtreelikelihood/ | 1 (0.1%) | 4 (0.0%) | 4.0 |
| src/dr/app/beauti/taxonsetspanel/ | 1 (0.1%) | 4 (0.0%) | 4.0 |
| src/dr/inference/parallel/ | 2 (0.1%) | 3 (0.0%) | 1.5 |
| src/dr/evomodel/arg/likelihood/ | 3 (0.2%) | 3 (0.0%) | 1.0 |
| src/dr/evolution/sequence/ | 2 (0.1%) | 3 (0.0%) | 1.5 |
| examples/calibrations/ | 6 (0.3%) | 3 (0.0%) | 0.5 |
| src/dr/evomodel/beagle/treelikelihood/ | 1 (0.1%) | 2 (0.0%) | 2.0 |
| src/dr/evomodel/alternativesplicing/ | 2 (0.1%) | 2 (0.0%) | 1.0 |
| src/dr/app/misc/ | 1 (0.1%) | 2 (0.0%) | 2.0 |
| / | 1 (0.1%) | 2 (0.0%) | 2.0 |
| src/dr/math/matrixAlgebra/ | 1 (0.1%) | 1 (0.0%) | 1.0 |
| src/dr/evomodelxml/branchratemodel/ | 1 (0.1%) | 1 (0.0%) | 1.0 |
| src/dr/app/beauti/treespanel/ | 1 (0.1%) | 1 (0.0%) | 1.0 |
| native/ | 2 (0.1%) | 1 (0.0%) | 0.5 |
| src/dr/app/beauti/traitspanel/ | 1 (0.1%) | 0 (0.0%) | 0.0 |
| lib/ | 2 (0.1%) | 0 (0.0%) | 0.0 |
Re-organize calibration code. add experimental "approximate" marginal.
405 lines of code changed in 7 files:
add some expectations
32 lines of code changed in 12 files:
add a few calibration test files
722 lines of code changed in 7 files:
add all worked out closed form calibration marginals: parent of clade, two nested clades, clade and root.
168 lines of code changed in 5 files:
Fix one bug in constrained generation of initial trees. bug was due to creation on 1 taxon tree with height 0.
7 lines of code changed in 2 files:
Correctly handle calibration on root
14 lines of code changed in 1 file:
validation for RPN expressions
57 lines of code changed in 6 files:
Add chs, log and exp tp RPN calculator
Add Tree height statistic parser tp .properties
Special calibration cases use RPN expression.
170 lines of code changed in 7 files:
First implementation of correct calibration density for one monophyletic clade inside a Yule tree.
Allow an "explicit" correction via pre computed coefficients, used when putting additional constraints on the topology
43 lines of code changed in 5 files:
First implementation of correct calibration density for one monophyletic clade inside a Yule tree.
The internal interface it a little clunky since the concept does not fit well with the current classes implementing the various speciation priors.
The XML interface is via an additional 'calibration' element inside
the speciationLikelihood. The element contains the taxa and a distribution for the root of the taxa. For a single taxon list, the distribution is for the parent of the taxon, which has no other constraints.
<speciationLikelihood id="speciationlike">
<model>
<yuleModel idref="yule"/>
</model>
<speciesTree>
<treeModel idref="treeModel"/>
</speciesTree>
<calibration>
<exponentialDistributionModel>
<mean>
<parameter id="mean" value="1"/>
</mean>
</exponentialDistributionModel>
<taxa>
<taxon idref="tip1"/>
<taxon idref="tip2"/>
</taxa>
</calibration>
</speciationLikelihood>
158 lines of code changed in 10 files:
automatic intelliJ minor syntax fixes
8 lines of code changed in 6 files:
Allows any number of tempratures in command line
92 lines of code changed in 2 files:
Give a more intuitive error message
5 lines of code changed in 1 file:
Give a sane message when no files are given (since files with a non-approved extension, even if explicitly given on the command line, are silently filtered out)
5 lines of code changed in 1 file:
Now that times() is used to print values in log, change implementation to return only the externally visible ones.
2 lines of code changed in 2 files:
Catch errors which lead to beast infinite loop or failure in parsers. more detailed error messages.
18 lines of code changed in 8 files:
Ignore nonesense ESS on constant values. integer ESS values in panel.
11 lines of code changed in 1 file:
Add an option to dump all demographics to a file
33 lines of code changed in 4 files:
Remove annoying debug print
3 lines of code changed in 1 file:
Allow multiple moves
16 lines of code changed in 1 file:
(423 more)