[root]/src/dr/evomodel/continuous
![]() plink
(1 files, 67 lines)
plink
(1 files, 67 lines)
| Author | Changes | Lines of Code | Lines per Change |
|---|---|---|---|
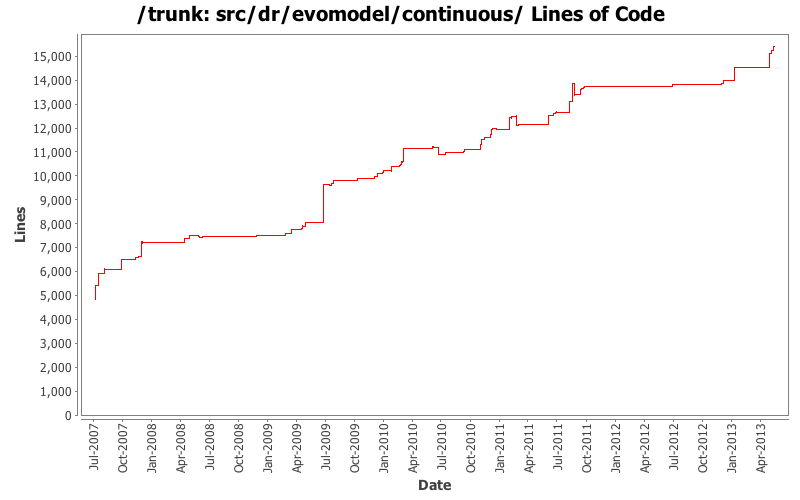
| Totals | 413 (100.0%) | 21545 (100.0%) | 52.1 |
| msuchard | 227 (55.0%) | 12448 (57.8%) | 54.8 |
| rambaut | 112 (27.1%) | 5479 (25.4%) | 48.9 |
| alexei.drummond | 39 (9.4%) | 2408 (11.2%) | 61.7 |
| gcybis@yahoo.com.br | 2 (0.5%) | 595 (2.8%) | 297.5 |
| jheled | 22 (5.3%) | 388 (1.8%) | 17.6 |
| mandevsgill | 5 (1.2%) | 165 (0.8%) | 33.0 |
| philippe.lemey@gmail.com | 4 (1.0%) | 57 (0.3%) | 14.2 |
| tbcbedford@gmail.com | 2 (0.5%) | 5 (0.0%) | 2.5 |
A few more minor changes (essentially all operations involving meanCache and storedMeanCache now done by cacheHelper). Rewritten code does not break anything.
0 lines of code changed in 2 files:
Some very minor changes
160 lines of code changed in 1 file:
Starting on diffusion with drift
5 lines of code changed in 2 files:
Fixing transformed trees when random
121 lines of code changed in 16 files:
Finally interfacing multivariate traits, although they are now generally integrated out.
46 lines of code changed in 4 files:
Latent Liability model
595 lines of code changed in 2 files:
Fixing jitter in IMTL
5 lines of code changed in 1 file:
Added unit-IntervalLatentLiabilityLikelihood for VNTRs; plan to refactor to remove duplicated code once more fully tested.
553 lines of code changed in 4 files:
Delegated handling of missing values in IntegratedMultivariateTraitLikelihood with a view towards enabling partially missing values
184 lines of code changed in 5 files:
Fixed zero-branch length bug in IntegratedMultivariateTraitLikelihood and harmonized behavior of NonPhylogeneticMultivariateTraitLikelihood for model selection.
53 lines of code changed in 3 files:
Checking equations for Pybus et al. manuscript
70 lines of code changed in 2 files:
Implementing transformed trees and Pagel's lambda estimator
29 lines of code changed in 1 file:
Fixed bug in calculation of log density in BiasedMultivariateDiffusionModel. Branches should properly scale with time now. This seems to be working.
5 lines of code changed in 2 files:
Can specify non-integer degrees-of-freedom for Wishart distributions
11 lines of code changed in 1 file:
Fixed degrees-of-freedom in Wishart statistics for NonPhylogeneticMultivariateTraitLikelihood
28 lines of code changed in 1 file:
Enabled Gibbs sampling of precision matrix in NonPhylogeneticMultivariateTraitLikelihood
69 lines of code changed in 2 files:
Fixed store/restore in WeightedMixtureModel
0 lines of code changed in 1 file:
Playing with rescaling for the non-phylogenetic model
27 lines of code changed in 1 file:
Implemented a null model to test for phylogenetic correlation of continuous traits
254 lines of code changed in 7 files:
Latent liability model appears to work well.
40 lines of code changed in 1 file:
Moved antigenic stuff into its own package.
0 lines of code changed in 3 files:
Implemented Joe Felsenstein's latent liability model with a single new class to BEAST.
243 lines of code changed in 3 files:
Fixed up a few things.
551 lines of code changed in 3 files:
Various bits
6 lines of code changed in 1 file:
Work on discrete Antigenic MDS
11 lines of code changed in 2 files:
Some refactoring of MatrixParameter (renamed getNumberOfParameters to getParameterCount). Also allowed MultivariateNormalPrior take a matrix parameter and assume the individual parameters within this represent independent draws from the MVN.
34 lines of code changed in 4 files:
Discrete AG model...
55 lines of code changed in 1 file:
First attempt at a discrete antigenic cluster classifier.
400 lines of code changed in 3 files:
16 lines of code changed in 2 files:
Improvements and tweaks to Antigenic MDS
5 lines of code changed in 1 file:
Improvements and tweaks to Antigenic MDS
102 lines of code changed in 2 files:
Seem to have got the Antigenic MDS working correctly with threshold assay values and replicate measurements.
136 lines of code changed in 2 files:
Slowly beating this code into shape
154 lines of code changed in 2 files:
Resuming work on this...
3 lines of code changed in 1 file:
uncommitting an erroneous revision
0 lines of code changed in 2 files:
Fixing some issues with row/column indexing in the sparse matrix
18 lines of code changed in 2 files:
Added skeleton class for non-phylogenetic trait model for model comparison
50 lines of code changed in 2 files:
Implementing paired virus-antiserum measurements (for when a titre is available for a serum raised against the virus being tested - this will be assume to be drawn from an identical distribution across all such pairs).
44 lines of code changed in 2 files:
Reads text based files which map repeated assays to antisera and viruses to corresponding antisera (these will be assumed to be drawn from an independent distribution independent of location).
103 lines of code changed in 1 file:
2 lines of code changed in 1 file:
(172 more)