

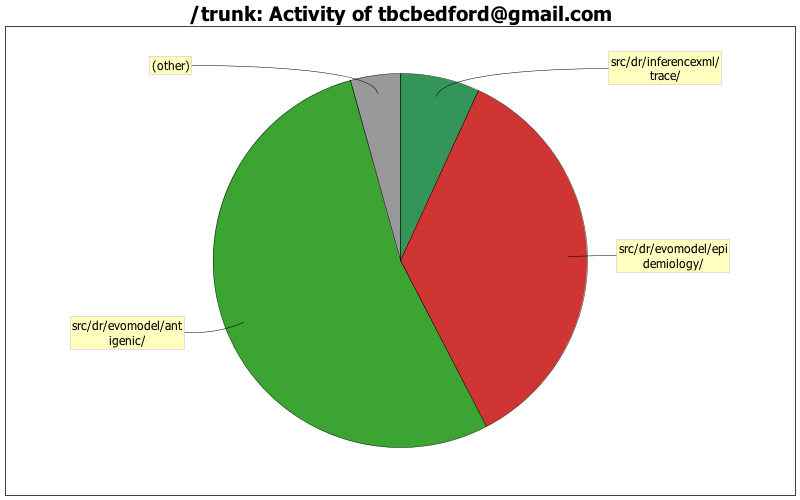
| Directory | Changes | Lines of Code | Lines per Change |
|---|---|---|---|
| Totals | 170 (100.0%) | 5831 (100.0%) | 34.3 |
| src/dr/evomodel/antigenic/ | 72 (42.4%) | 3114 (53.4%) | 43.2 |
| src/dr/evomodel/epidemiology/ | 51 (30.0%) | 2067 (35.4%) | 40.5 |
| src/dr/inferencexml/trace/ | 9 (5.3%) | 402 (6.9%) | 44.6 |
| src/dr/app/tracer/analysis/ | 11 (6.5%) | 87 (1.5%) | 7.9 |
| src/dr/evomodel/branchratemodel/ | 3 (1.8%) | 63 (1.1%) | 21.0 |
| src/dr/inference/trace/ | 5 (2.9%) | 62 (1.1%) | 12.4 |
| src/dr/app/beast/ | 10 (5.9%) | 13 (0.2%) | 1.3 |
| src/dr/app/tracer/application/ | 3 (1.8%) | 11 (0.2%) | 3.6 |
| src/dr/evomodel/continuous/ | 2 (1.2%) | 5 (0.1%) | 2.5 |
| src/dr/app/gui/chart/ | 3 (1.8%) | 4 (0.1%) | 1.3 |
| src/dr/math/distributions/ | 1 (0.6%) | 3 (0.1%) | 3.0 |
Small correction, not working yet.
5 lines of code changed in 2 files:
My attempt to get discreteTraitBranchRateModel working. It's almost there. However, the function:
int state = ((int[])trait.getTrait(tree, node))[traitIndex];
on line 386 is causing problems.
If getProcessValues is forced to an array of {1.0, 1.0} in getRawBranchRate then the MCMC proceeds as expected. However, if the return value of getProcessValues is set to {1.0, 1.0} then the problem persists.
58 lines of code changed in 1 file:
Ignore this. Testing homebrew upgrade mechanisms.
1 lines of code changed in 2 files:
Change "mergeColumns" to "mergeSerumIsolates" to agree with the rest of the spec.
6 lines of code changed in 2 files:
Fix bug in indexing. Proper number of columns now being recorded.
1 lines of code changed in 1 file:
Finalizing schema to agree with paper. Row effects -> Virus effects. Column effects -> Serum effects. Also, reordering input file to be: virus isolate, virus strain, virus year, serum isolate, serum strain, serum year, titre.
23 lines of code changed in 1 file:
This is a refactor of AntigenicLikelihood. It's been slimmed down substantially. I've tried to cut down on the all the redirection involved in setting up parameters. There is now just equal-sized vectors of names, locations, offsets and effects for both viruses and sera. "Column effects" and locations now have to refer to the same set of entities, unlike previously where locations could refer to strains and effects could refer to isolates.
This is currently broken in some way and giving "SEVERE: State was not correctly calculated after an operator move." errors.
781 lines of code changed in 1 file:
Change AntigenicDistancePrior to take new Offsets parameter (rather than Dates parameter) and remove deprecated AntigenicJumpPrior.
216 lines of code changed in 4 files:
Include drifted traits logger in dev parsers.
1 lines of code changed in 1 file:
Implement logging of drifted tree traits.
155 lines of code changed in 2 files:
Improving caching by tracking column effect and row effect updates.
24 lines of code changed in 1 file:
Small change to parameter syntax (virusOffset to virusOffsets).
30 lines of code changed in 2 files:
Include DriftedLocationStatistic in dev parsers.
1 lines of code changed in 1 file:
Implement statistic that records drifted locations. This required storing virusDates and serumDates parameters from AntigenicLikelihood and passing these, plus the locationDriftParameter, to the new DriftedLocationsStatistic.
162 lines of code changed in 2 files:
Depecrate antigenic drift priors. Now incorporated directly into antigenicLikelihood.
49 lines of code changed in 4 files:
Streamline antigenic likelihood a bit, removing virusDates parameter.
19 lines of code changed in 1 file:
Reinstate linear initial conditions.
4 lines of code changed in 1 file:
Fix bug in assigning dates.
10 lines of code changed in 1 file:
Implement antigenic drift on map distances.
48 lines of code changed in 1 file:
Include antigenic drift parameter. Doesn't do anything yet.
20 lines of code changed in 1 file:
(71 more)