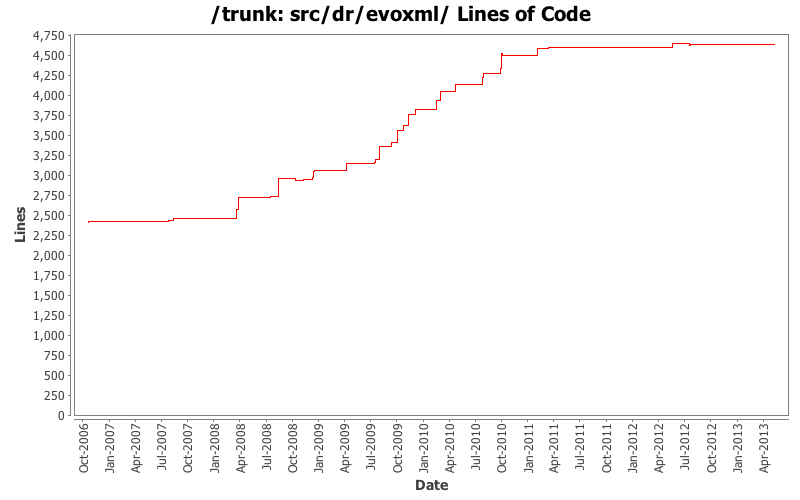
[root]/src/dr/evoxml
![]() util
(4 files, 400 lines)
util
(4 files, 400 lines)
| Author | Changes | Lines of Code | Lines per Change |
|---|---|---|---|
| Totals | 162 (100.0%) | 2965 (100.0%) | 18.3 |
| alexei.drummond | 27 (16.7%) | 648 (21.9%) | 24.0 |
| rambaut | 35 (21.6%) | 637 (21.5%) | 18.2 |
| msuchard | 17 (10.5%) | 466 (15.7%) | 27.4 |
| akaruiws | 10 (6.2%) | 373 (12.6%) | 37.3 |
| dong.w.xie | 28 (17.3%) | 328 (11.1%) | 11.7 |
| alexander.alekseyenko | 3 (1.9%) | 225 (7.6%) | 75.0 |
| aaron.darling | 3 (1.9%) | 187 (6.3%) | 62.3 |
| jheled | 37 (22.8%) | 100 (3.4%) | 2.7 |
| fbielejec | 1 (0.6%) | 1 (0.0%) | 1.0 |
| Michael.DefoinPLatel | 1 (0.6%) | 0 (0.0%) | 0.0 |
Added parsing and setting of date precisions in BEAUti plus generation of uniform sampling within these bounds. Also fixed the date calculations so Jan 1st (say 2013) is 2013.0.
1 lines of code changed in 2 files:
Porting fixes for issue http://code.google.com/p/beast-mcmc/issues/detail?id=671 to trunk.
7 lines of code changed in 2 files:
Precision field in date now seems to work correctly: if you give a precision="1.0" to a date then it assumes that the date was sampled from the given t to t+1 in whatever units are being used (forwards and backwards dates are accommodated). If a treemodel leafHeight is created then this range is automatically added as a bound allowing a randomwalk operator to be used.
2 lines of code changed in 1 file:
Overhauled the date system so that the 'height' timescale is now universal to all taxa. Previously there was an issue if you create independent trees on different taxon sets, they would have their own 'zero' point depending on their most recently sampled taxon. Now, when you call 'setDate' for a taxon (as done by the TaxonParser) it keeps track of the globally most recent sample (in a static variable) and can give the global height from a given date object using a static function. Taxon objects also have a 'getHeight' methods that uses this system. The two main sources of trees (CoalescentSimulator and NewickParser) now use these heights (simplifying their own code). Runs benchmarks correctly but keep an eye out for issues.
9 lines of code changed in 3 files:
Imported patch attachd to issue 430 - a PearsonCorrelation statistic.
56 lines of code changed in 1 file:
Fixed an issue in BEAUti with robust counting XML generated where completely gapped sites were being stripped (these are sometimes used to maintain reading frame).
1 lines of code changed in 2 files:
Restored (but renamed to improve clarity) the UI to enable state counting other than the dNdS Robust Counting.
0 lines of code changed in 1 file:
added merge patterns tag
1 lines of code changed in 1 file:
trunk:solve Issue 550: MicroSat pattern does not generate properly
3 lines of code changed in 2 files:
Trunk: continue to work on BEAUti microsat, but writing different taxa function still has a bug.
2 lines of code changed in 1 file:
Trunk: more work on micro-sat data in BEAUti.
10 lines of code changed in 1 file:
Fixed a parser that allows you to create patterns of constant sites which can then me merged with polymorphic sites to reconstruct a full pattern set.
3 lines of code changed in 4 files:
Added a parser that allows you to create patterns of constant sites which can then me merged with polymorphic sites to reconstruct a full pattern set.
93 lines of code changed in 1 file:
Trunk: @Override is JDK1.6 code.
15 lines of code changed in 6 files:
Adding a new model of hidden linkage among reads in a metagenomic dataset. The model and its application to metagenomic datasets is described in a manuscript in preparation by Aaron Darling and Jonathan Eisen. This is commit phase 1: adding new files. Phase 2 comes next, committing related changes to existing files.
187 lines of code changed in 3 files:
Getting aggressive with fixing zero-branch-lengths in starting trees; need comparison of why MutableTree.Utils is not working
59 lines of code changed in 2 files:
Fixed issue where pressing "Guess Traits" to create a new trait resulted in the Guess Traits Dialog appearing twice.
1 lines of code changed in 2 files:
Added a check for AttributePatternsParser that the attribute actually exists in at least one taxon.
8 lines of code changed in 2 files:
Classes for modelling the relative rates of loci.
49 lines of code changed in 1 file:
Add ability to select a random subset of taxa for analysis
82 lines of code changed in 1 file:
Bifractional diffusion is a pain
2 lines of code changed in 2 files:
Fixed bug in ascertainment correction (miscounted) and implemented in BEAGLE. Genomic-wide SNPs here we come!
7 lines of code changed in 2 files:
More work on the hypermutation model.
5 lines of code changed in 2 files:
Created an HypermutantAlignment that flags hypermutation contexts for use in the APOBECErrorModel. It does this by replacing 'A's by an ambiguity code for A/G if it is in the correct context in the alignment. These can then be compressed into unique site patterns so that speed is not affected.
Extracted a common abstract base class with ConvertAlignment which wraps an alignment.
86 lines of code changed in 1 file:
TRUNK: BEAUti start generator for location subst model and general subst model.
2 lines of code changed in 1 file:
Trunk refactoring: organize Data Tpye parser (split parsers).
106 lines of code changed in 1 file:
Allow parser options to *not* condense site patterns into a unique list; this is necessary for robust counting of S and N mutations
9 lines of code changed in 1 file:
Trunk refactoring: organize tree and treelikelihood parsers.
1 lines of code changed in 1 file:
Trunk refactoring: make evoxml parsers more organized.
11 lines of code changed in 12 files:
Trunk refactoring: RandomTaxaSampleParser.
120 lines of code changed in 1 file:
Refactoring geo-specific functionality into dr.geo (step 1). Also, adding in shell classes for many different spatial distributions
1 lines of code changed in 2 files:
Started work on bifractional continuous random walk models
55 lines of code changed in 1 file:
134 lines of code changed in 3 files:
code police
7 lines of code changed in 1 file:
added parsers for Lewis' Mk model with totaly ordered/unordered/custom ordered states.
64 lines of code changed in 1 file:
Calculates overall mean pairwise differences across multiple loci.
66 lines of code changed in 1 file:
Parser for microsatellite simulator.
80 lines of code changed in 1 file:
Comments changed, new rules added. Printing details of microsatellite at the start becomes optional.
19 lines of code changed in 2 files:
Changed comments.
1 lines of code changed in 1 file:
BEAUTi: keep reducing ModelOptions by create enum DateUnitsType, and fix the bug of r2223.
58 lines of code changed in 2 files:
(51 more)