

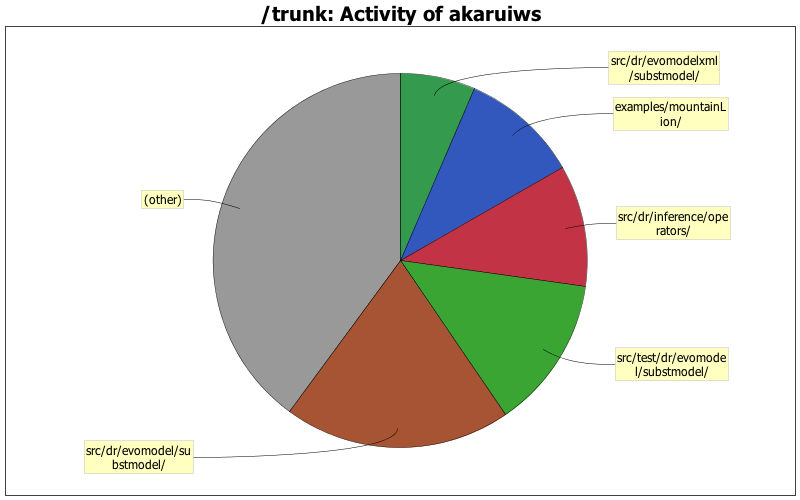
| Directory | Changes | Lines of Code | Lines per Change |
|---|---|---|---|
| Totals | 227 (100.0%) | 10509 (100.0%) | 46.2 |
| src/dr/evomodel/substmodel/ | 55 (24.2%) | 2064 (19.6%) | 37.5 |
| src/test/dr/evomodel/substmodel/ | 26 (11.5%) | 1387 (13.2%) | 53.3 |
| src/dr/inference/operators/ | 18 (7.9%) | 1105 (10.5%) | 61.3 |
| examples/mountainLion/ | 2 (0.9%) | 1072 (10.2%) | 536.0 |
| src/dr/evomodelxml/substmodel/ | 9 (4.0%) | 685 (6.5%) | 76.1 |
| src/dr/evomodel/sitemodel/ | 8 (3.5%) | 490 (4.7%) | 61.2 |
| examples/moa/ | 2 (0.9%) | 377 (3.6%) | 188.5 |
| src/dr/evoxml/ | 10 (4.4%) | 373 (3.5%) | 37.3 |
| src/dr/inferencexml/operators/ | 11 (4.8%) | 369 (3.5%) | 33.5 |
| src/dr/evomodel/treelikelihood/ | 5 (2.2%) | 313 (3.0%) | 62.6 |
| src/dr/evomodelxml/ | 9 (4.0%) | 309 (2.9%) | 34.3 |
| src/dr/evolution/datatype/ | 3 (1.3%) | 266 (2.5%) | 88.6 |
| src/test/dr/evomodel/sitemodel/ | 1 (0.4%) | 262 (2.5%) | 262.0 |
| src/dr/evomodel/tree/ | 6 (2.6%) | 257 (2.4%) | 42.8 |
| src/dr/evomodelxml/sitemodel/ | 5 (2.2%) | 246 (2.3%) | 49.2 |
| src/dr/app/seqgen/ | 10 (4.4%) | 242 (2.3%) | 24.2 |
| src/dr/inferencexml/distribution/ | 2 (0.9%) | 138 (1.3%) | 69.0 |
| src/dr/app/beast/ | 14 (6.2%) | 128 (1.2%) | 9.1 |
| src/dr/inference/distribution/ | 3 (1.3%) | 108 (1.0%) | 36.0 |
| src/dr/inference/model/ | 6 (2.6%) | 100 (1.0%) | 16.6 |
| src/dr/inferencexml/model/ | 1 (0.4%) | 67 (0.6%) | 67.0 |
| src/dr/evolution/alignment/ | 1 (0.4%) | 50 (0.5%) | 50.0 |
| src/dr/evolution/distance/ | 1 (0.4%) | 35 (0.3%) | 35.0 |
| src/dr/evomodelxml/treelikelihood/ | 1 (0.4%) | 25 (0.2%) | 25.0 |
| src/dr/xml/ | 2 (0.9%) | 23 (0.2%) | 11.5 |
| src/dr/math/ | 4 (1.8%) | 5 (0.0%) | 1.2 |
| / | 2 (0.9%) | 5 (0.0%) | 2.5 |
| examples/Phylogeography/continuous/ | 2 (0.9%) | 4 (0.0%) | 2.0 |
| src/dr/app/beauti/options/ | 3 (1.3%) | 2 (0.0%) | 0.6 |
| src/dr/math/distributions/ | 1 (0.4%) | 1 (0.0%) | 1.0 |
| examples/release/ | 1 (0.4%) | 1 (0.0%) | 1.0 |
| examples/ | 1 (0.4%) | 0 (0.0%) | 0.0 |
| doc/tutorial/DivergenceDating/1.2_Primates/latex/figures/ | 2 (0.9%) | 0 (0.0%) | 0.0 |
Missed correction.
17 lines of code changed in 2 files:
Modify to handle ambiguity.
3 lines of code changed in 2 files:
Provide more realistic default value for model parameter just like providing more realistic starting tree to the analysis to prevent numerical instability or whatever silly stuff that does not allow the analysis to start running.
1 lines of code changed in 2 files:
0 lines of code changed in 2 files:
Change the performance suggestion.
5 lines of code changed in 1 file:
Correct tag name.
2 lines of code changed in 2 files:
Tidy up more msatBMA related things.
73 lines of code changed in 6 files:
Correcting according to changes made to the models.
3 lines of code changed in 2 files:
Tidy up MsatBMA related things.
739 lines of code changed in 13 files:
Create set methods for max and min.
8 lines of code changed in 1 file:
Deleting the accidently committed file.
0 lines of code changed in 1 file:
BMA for nucleotide substition models and gamma site model.
1092 lines of code changed in 12 files:
GTR examples added.
59 lines of code changed in 1 file:
TN93 example added.
11 lines of code changed in 1 file:
Some quick JUnit tests for NtdBMA model to check whether there are any major problems. More useful tests yet to come.
227 lines of code changed in 1 file:
Handle the variable change more efficiently.
27 lines of code changed in 1 file:
Model averaging for nucleotide models.
157 lines of code changed in 3 files:
add new constructor.
18 lines of code changed in 2 files:
updates for watkins msat model classes.
117 lines of code changed in 3 files:
Simulator for geographic locations by homogeneous diffusion.
121 lines of code changed in 3 files:
(87 more)