

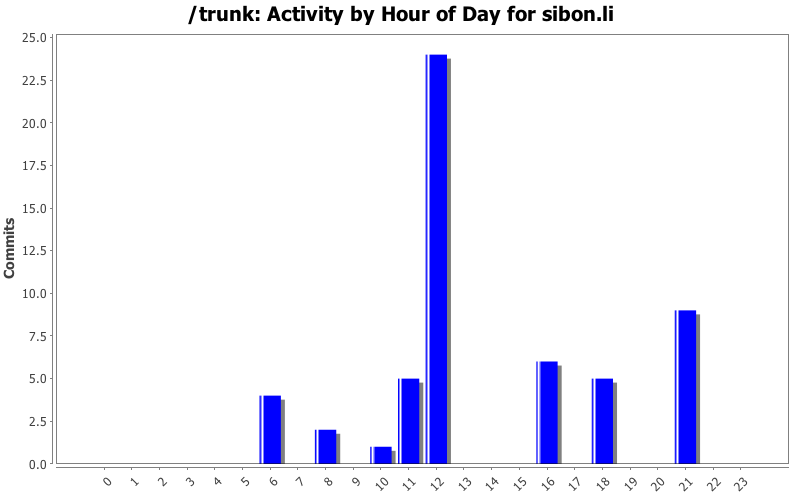
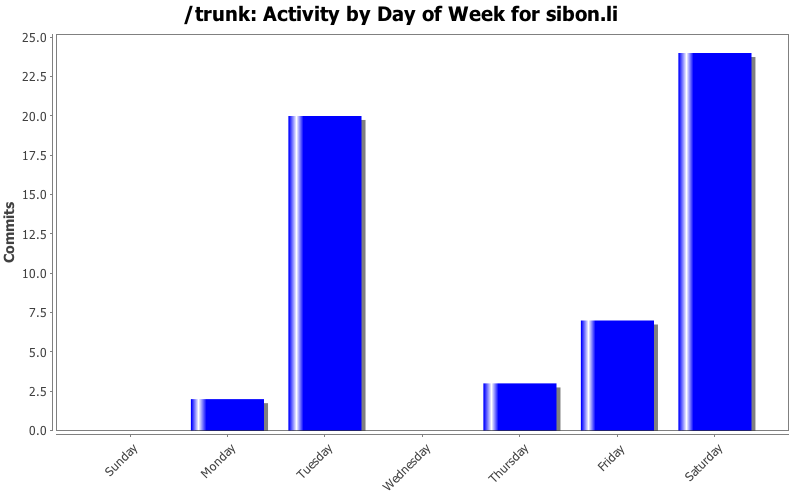
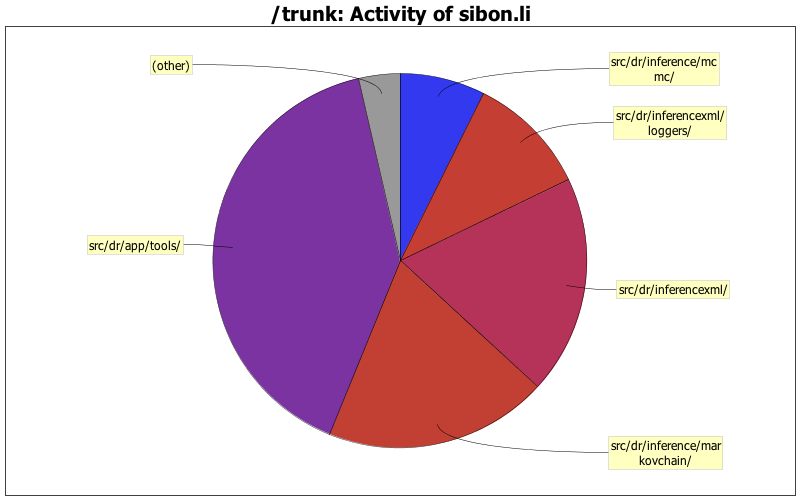
| Directory | Changes | Lines of Code | Lines per Change |
|---|---|---|---|
| Totals | 56 (100.0%) | 6336 (100.0%) | 113.1 |
| src/dr/app/tools/ | 14 (25.0%) | 2555 (40.3%) | 182.5 |
| src/dr/inference/markovchain/ | 5 (8.9%) | 1223 (19.3%) | 244.6 |
| src/dr/inferencexml/ | 6 (10.7%) | 1197 (18.9%) | 199.5 |
| src/dr/inferencexml/loggers/ | 4 (7.1%) | 668 (10.5%) | 167.0 |
| src/dr/inference/mcmc/ | 4 (7.1%) | 467 (7.4%) | 116.7 |
| src/test/dr/evomodel/substmodel/ | 2 (3.6%) | 178 (2.8%) | 89.0 |
| src/dr/inference/distribution/ | 2 (3.6%) | 22 (0.3%) | 11.0 |
| src/dr/inferencexml/distribution/ | 2 (3.6%) | 9 (0.1%) | 4.5 |
| src/dr/evomodel/treelikelihood/ | 2 (3.6%) | 5 (0.1%) | 2.5 |
| / | 5 (8.9%) | 5 (0.1%) | 1.0 |
| src/dr/evolution/alignment/ | 1 (1.8%) | 4 (0.1%) | 4.0 |
| src/test/dr/evomodel/branchratemodel/ | 2 (3.6%) | 1 (0.0%) | 0.5 |
| src/dr/evomodel/branchratemodel/ | 2 (3.6%) | 1 (0.0%) | 0.5 |
| src/dr/app/beagle/evomodel/treelikelihood/ | 2 (3.6%) | 1 (0.0%) | 0.5 |
| lib/ | 3 (5.4%) | 0 (0.0%) | 0.0 |
Deleted a non-essential tool that I wrote.
0 lines of code changed in 1 file:
Fixed AncestralSequenceAnnotator to use KALIGN instead of CLUSTAL (much faster)
17 lines of code changed in 1 file:
Added an interface for delegating tasks in the Markov Chain
143 lines of code changed in 4 files:
Commented out all of my last commit until issues resolved
1202 lines of code changed in 9 files:
Test build
1 lines of code changed in 1 file:
Added blue-beast.jar and modified build xmls
3 lines of code changed in 4 files:
Added classes to enable implicit convergence assessment checks using BLUE
1255 lines of code changed in 4 files:
Changes to encapsulation in MCMC.java and MarkovChain.java. MarkovChain.java no longer final and some private variables changed to protected
33 lines of code changed in 2 files:
Correction, parsers pending updates to other files. Currently commented out
791 lines of code changed in 4 files:
Added two new parsers for BLUE integration
467 lines of code changed in 2 files:
Improved presentation for AncestralSequenceAnnotator and GetAncestralSequenceFromSplitTrait
14 lines of code changed in 3 files:
Fixed a bug that made AncestralSequenceAnnotator not work properly for BAli-Phy files. Now provided it an option to take in BEAST input tree files
498 lines of code changed in 1 file:
Work in progress on AncestralSequenceAnnotator, will be fixed by end of today
239 lines of code changed in 1 file:
Created an app - GetAncestralSequenceFromSplitTrait, which does just that.
347 lines of code changed in 3 files:
Fixing an error in my last commit
0 lines of code changed in 1 file:
Transferring my changes from branch 1.6 to trunk:
Changes to LogNormalDistributionModel to allow for the stdev value to also not
be in log space (r3774)
Changes to UncorrelatedRelaxedClockTest due to changes in
LogNormalDistributionModel in r3774 (r3775)
Minor change in MixtureModelBranchRates to fix an issue caused by changes in
revision 3735 (r3773)
Bug fix for AncestralSequenceAnnotator plus added a window to select
input/output files (r3819)
Fixed spelling for GeneralSubstitutionModelTest (r3833)
Fixed a RuntimeException for AncestralStateTreeLikelihood (r3957)
217 lines of code changed in 13 files:
Major work on AncestralSequenceAnnotator. Mostly generalization for different models and datatypes. Added MakeAncestralSequenceAnnotatedTree, which takes BAli-Phy output and turns it into a BEAST compatible format
1109 lines of code changed in 2 files: