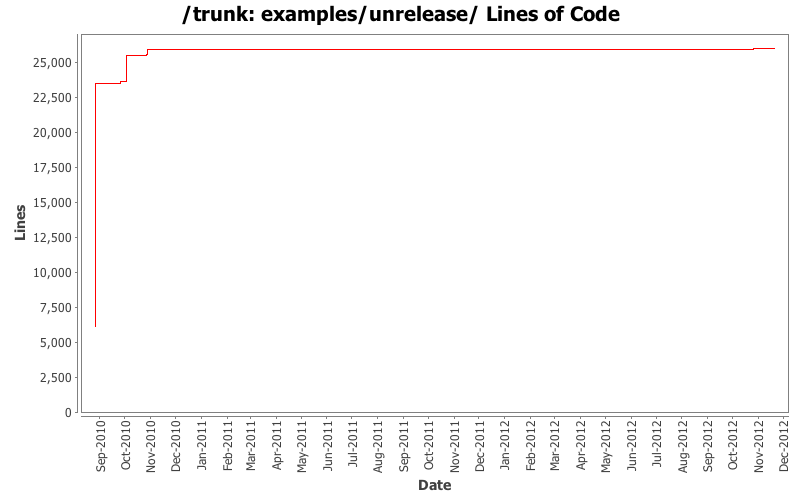
-
alexander.alekseyenko
2012-11-20 09:35
Rev.: 5244
-
7 lines of code changed in 2 files:
-
examples/unrelease:
testMVPolyaLikelihood2.xml (+7 -2)
-
alexander.alekseyenko
2012-11-17 11:24
Rev.: 5238
-
26 lines of code changed in 1 file:
-
examples/unrelease:
testMVPolyaLikelihood2.xml (+26 -6)
-
alexander.alekseyenko
2012-10-26 09:33
Rev.: 5209
-
86 lines of code changed in 1 file:
-
examples/unrelease:
testMVPolyaLikelihood2.xml (+86)
-
msuchard
2010-10-28 03:38
Rev.: 3685
-
334 lines of code changed in 2 files:
-
examples/unrelease:
testBranchSiteModel.xml (del),
testBranchSubstitutionModel.xml (new 334)
-
msuchard
2010-10-27 12:02
Rev.: 3683
-
91 lines of code changed in 2 files:
-
examples/unrelease:
testBranchSiteModel.xml (new),
testIntegratedConstantCoalescent.xml (new 91)
-
aaron.darling
2010-10-02 15:30
Rev.: 3595
-
1880 lines of code changed in 1 file:
-
examples/unrelease:
bushSausageUnknown.xml (new 1880)
-
alexander.alekseyenko
2010-09-25 04:58
Rev.: 3571
-
93 lines of code changed in 1 file:
-
examples/unrelease:
testMVPolyaLikelihood.xml (new 93)
-
alexander.alekseyenko
2010-09-24 04:20
Rev.: 3567
-
2 lines of code changed in 2 files:
-
examples/unrelease:
testIBDweights.xml (new),
testPathSampling.xml (new 2)
-
dong.w.xie
2010-08-27 16:15
Rev.: 3467
-
0 lines of code changed in 2 files:
-
examples/unrelease:
Msat (del),
Phylogeography (del)
-
dong.w.xie
2010-08-27 16:03
Rev.: 3465
-
0 lines of code changed in 25 files:
-
examples/unrelease:
testAlignmentStatistics.xml (del),
testAutocorrelatedRelaxedClock.xml (del),
testBinaryDollo.xml (del),
testCTMCMap.xml (del),
testCompleteHistorySimulator.xml (del),
testConjugateTraitLikelihood.xml (del),
testDiscreteMapDiffusion.xml (del),
testIntegratedTraitLikelihood.xml (del),
testLewisMk.xml (del),
testLinearRegression.xml (del),
testMLCalculator.xml (del),
testMSSD.xml (del),
testMarkovJumps.xml (del),
testMissingIntegratedTraitLikelihood.xml (del),
testMultivariateComparativeMethod.xml (del),
testNewLikelihood.xml (del),
testOTFPCLikelihood.xml (del),
testPopTreeModel.xml (del),
testRestrictedAncestralReconstruction.xml (del),
testRobustCounting.xml (del),
testSimplePathSampling.xml (del),
testTransmission.xml (del),
testTreePathSampling.xml (del),
testUniformizedMarkovJumps.xml (del),
testYuleOneSite.xml (del)
-
dong.w.xie
2010-08-27 13:28
Rev.: 3463
-
0 lines of code changed in 35 files:
-
examples/unrelease:
Mdor phygeo ages1.xml (del),
arg-ecoli.xml (del),
arg-lepto.xml (del),
arg-rokas.xml (del),
benchmark.xml (del),
benchmark2.xml (del),
demoCodonHistorySimulator.xml (del),
new_benchmark.xml (del),
testAncestralReconstruction.xml (del),
testAscertainment.xml (del),
testBSP.xml (del),
testCodonLikelihood.xml (del),
testComparativeMethod.xml (del),
testCovarionHKY.xml (del),
testGeneralSubstitutionModel.xml (del),
testIA.xml (del),
testIBDweights.xml (del),
testLikelihood.xml (del),
testLocalClock.xml (del),
testLognormalPrior.xml (del),
testMC3.xml (del),
testMCMC.xml (del),
testML.xml (del),
testMVOU.xml (del),
testMixedModel.xml (del),
testMultiThreading.xml (del),
testNormalModelGibbs.xml (del),
testPMD.xml (del),
testPathSampling.xml (del),
testPosteriorIBD.xml (del),
testRandomLocalClock.xml (del),
testSequenceSimulator.xml (del),
testStrictClock.xml (del),
testTransmissionStatistic.xml (del),
testUncorrelatedRelaxedClock.xml (del)
-
dong.w.xie
2010-08-27 11:20
Rev.: 3462
-
0 lines of code changed in 1 file:
-
examples/unrelease:
calibrations (del)
-
dong.w.xie
2010-08-27 11:12
Rev.: 3460
-
0 lines of code changed in 1 file:
-
examples/unrelease:
treePriors (del)
-
dong.w.xie
2010-08-26 16:43
Rev.: 3459
-
0 lines of code changed in 1 file:
-
examples/unrelease:
Data (del)
-
dong.w.xie
2010-08-26 16:11
-
17429 lines of code changed in 65 files:
-
examples/unrelease:
Data (new),
Mdor phygeo ages1.xml (new 1475),
Msat (new),
Phylogeography (new),
arg-ecoli.xml (new),
arg-lepto.xml (new),
arg-rokas.xml (new),
benchmark.xml (new),
benchmark2.xml (new),
calibrations (new),
demoCodonHistorySimulator.xml (new 88),
new_benchmark.xml (new 371),
testAlignmentStatistics.xml (new 23),
testAncestralReconstruction.xml (new 245),
testAscertainment.xml (new 174),
testAutocorrelatedRelaxedClock.xml (new 460),
testBSP.xml (new 324),
testBinaryDollo.xml (new 148),
testCTMCMap.xml (new 312),
testCodonLikelihood.xml (new 1183),
testComparativeMethod.xml (new 292),
testCompleteHistorySimulator.xml (new 200),
testConjugateTraitLikelihood.xml (new 124),
testCovarionHKY.xml (new 339),
testDiscreteMapDiffusion.xml (new 112),
testGeneralSubstitutionModel.xml (new 183),
testIA.xml (new 120),
testIBDweights.xml (new 166),
testIntegratedTraitLikelihood.xml (new 115),
testLewisMk.xml (new 31),
testLikelihood.xml (new 727),
testLinearRegression.xml (new 126),
testLocalClock.xml (new 413),
testLognormalPrior.xml (new 62),
testMC3.xml (new 300),
testMCMC.xml (new 175),
testML.xml (new 165),
testMLCalculator.xml (new 6),
testMSSD.xml (new 203),
testMVOU.xml (new 302),
testMarkovJumps.xml (new 150),
testMissingIntegratedTraitLikelihood.xml (new 115),
testMixedModel.xml (new 148),
testMultiThreading.xml (new 423),
testMultivariateComparativeMethod.xml (new 310),
testNewLikelihood.xml (new 477),
testNormalModelGibbs.xml (new 109),
testOTFPCLikelihood.xml (new 477),
testPMD.xml (new 1550),
testPathSampling.xml (new 236),
testPopTreeModel.xml (new 39),
testPosteriorIBD.xml (new 194),
testRandomLocalClock.xml (new 321),
testRestrictedAncestralReconstruction.xml (new 255),
testRobustCounting.xml (new 452),
testSequenceSimulator.xml (new 143),
testSimplePathSampling.xml (new 251),
testStrictClock.xml (new 334),
testTransmission.xml (new 742),
testTransmissionStatistic.xml (new 682),
testTreePathSampling.xml (new 262),
testUncorrelatedRelaxedClock.xml (new 418),
testUniformizedMarkovJumps.xml (new 152),
testYuleOneSite.xml (new 225),
treePriors (new)